Hereditary Cancer Panel Positive Control
This Oxford Nanopore Technologies dataset release includes positive control data from a single PromethION Flow Cell for three barcoded samples: Two barcodes correspond to sequencing runs of the NIBSC (code: 11/218 CE-IVD (Discontinued)), which contains two deletions spanning 2 and 6 exons in the MSH2 gene. A third barcode corresponds to a run of the LGC/SeraCare Seraseq, which includes BRCA1 and BRCA2 deletions of 1 and 5 exons, respectively. The data was generated and processed using the Hereditary Cancer Panel (HCP) workflow.
BRCA1/2 exon deletions are correctly identified with the LGC/SeraCare Seraseq BRCA 1/2 Exon Deletions DNA Mix. MSH2 deletions are correctly identified using the NIBSC MLH1/MSH2 Exon Copy Number Reference Panel.
Sample
| Detail | Description |
|---|---|
| Sample Name | NIBSC MLH1/MSH2 (MSH2 deletion exons 1-6), NIBSC MLH1/MSH2 (MSH2 deletion exons 1-2), Seraseq (730-0570) |
| Organism | Human |
| Molecule Type | gDNA |
| Sample Type | DNA standard |
| Biological replicates | 1 |
| Flow Cell replicates | 1 |
| Hereditary Cancer Panel | HCP18 (18 samples) | HCP72 (72 samples) |
| Sample Source | NIBSC (x2) and LGC/SeraCare Seraseq |
Preparation
Sample preparation was performed according to protocols contained within the Hereditary Cancer Panel Bundle.
| Detail | Description |
|---|---|
| Extraction and Library Prep | Hereditary Cancer Panel |
| Kit | Native Barcode ligation Kit V14 (SQK-NBD114-24) |
Further preparation information such as sample storage suggestions can be found on the Oxford Nanopore Website.
Sequencing
Sequence data were generated using the following configuration:
| Attribute | Value |
|---|---|
| Flow Cell | FLO-PRO114M |
| Device | PromethION |
| Chemistry | R10.4.1 |
| Basecall Model | dna_r10.4.1_e8.2_400bps_hac@v4.3.0 |
| Mod Model | 5mC & 5hmC CG contexts |
| MinKNOW Version | MinKnow 6.4 (25.03.7 focal) |
Data Download
The dataset is available for anonymous download, without login, from a public Amazon Web Services S3 bucket. The bucket is part of the Open Data on AWS project enabling sharing and analysis of a wide range of data. The data can be downloaded with the AWS CLI command:
aws s3 sync --no-sign-request s3://ont-open-data/hereditary_cancer_positive_control_2025.11 hereditary_cancer_positive_control_2025.11
See the tutorials page for information on downloading the dataset.
You can also browse and download the files in your web browser courtesy of 42basepairs.
| Folder name | Size | Description |
|---|---|---|
| raw | 730 GB | POD5 files |
| basecalls | 322 GB | BAM files |
| analysis | 91 GB | Workflow outputs |
Analysis
Data was analysed using the wf-hereditary-cancer workflow, an EPI2ME pipeline available as part of the Hereditary Cancer Panel Bundle. The analysis results are located in the S3 bucket under the prefix:
s3://ont-open-data/hereditary_cancer_positive_control_2025.11/analysis
BRCA1 and BRCA2 Structural Variant Detection (Seraseq)
To assess the workflow’s ability to detect structural variants in BRCA1 and BRCA2, the LGC/SeraCare Seraseq BRCA 1/2 Exon Deletions DNA Mix was used.
This reference sample, derived from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research: NA24385 cell line, includes engineered BRCA1/2 deletions at 60% VAF and 2× coverage relative to the background genome.
Samples were processed using the Hereditary Cancer Panel protocol (1 µg input DNA) with barcode 15. As the sequencing coverage was twice the target minimum of 30×, the data was in silico downsampled to 30× in duplicate replicates and analyzed using the wf-hereditary-cancer workflow.
Both BRCA1 and BRCA2 deletions were correctly identified in replicates (Table 1).
Table 1. Detection of BRCA1/2 Structural Variants in Seraseq BRCA 1/2 Exon Deletions DNA Mix
All variants were correctly identified by the workflow.
| Sample | Gene | Variant Type | Variant Length (bp) | Chr | Position | Expected VAF | Rep1 VAF | Rep2 VAF |
|---|---|---|---|---|---|---|---|---|
| Seraseq | BRCA1 | DEL | 3,571 | 17 | 43,091,345 | 60.1 | 48.28 | 52.27 |
| Seraseq | BRCA2 | DEL | 23,260 | 13 | 32,332,174 | 59.7 | 67.35 | 62.22 |
Support for these structural variants identified by the workflow can be visualised in IGV:
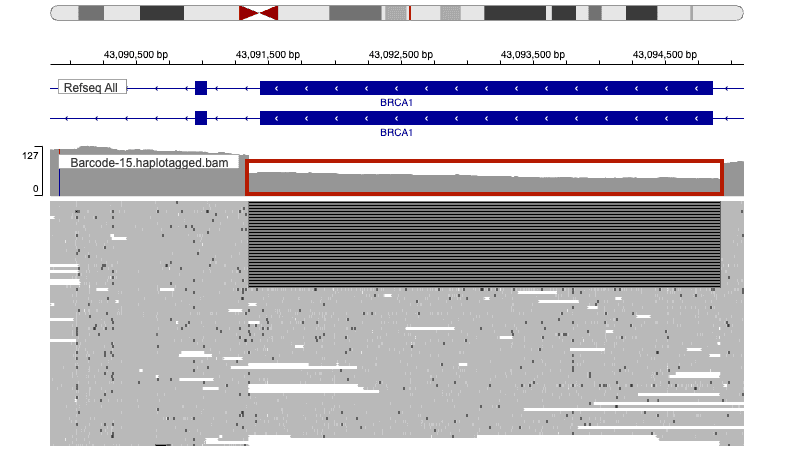
- BRCA1 (3.6 kb deletion): A large proportion of reads spanned the 3.6 kb deletion region, consistent with the expected c.671-56_4096+89del (p.Ala224_Leu1365del).
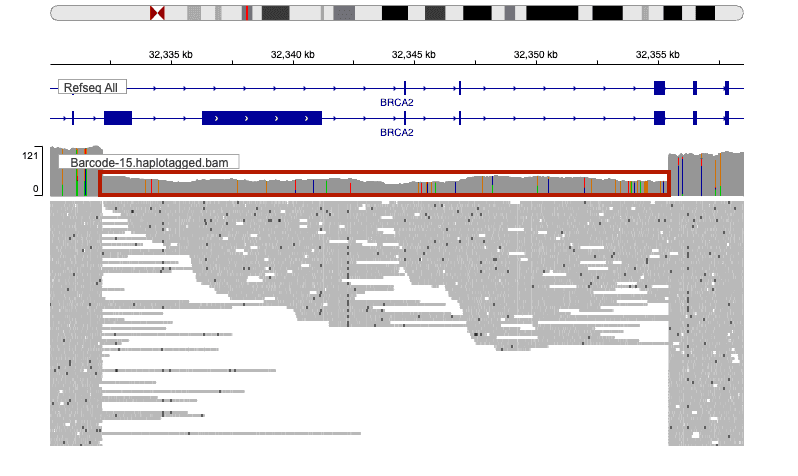
- BRCA2 (23 kb deletion): Soft-clipped reads were visible at the BRCA2 gene deletion boundaries, matching the expected c.795-97_7435+146del (p.Gly265_Leu2478del).
Figures 1 and 2 show representative IGV snapshots for each event.
MSH2 Structural Variant Detection (NIBSC Reference Panel)
The NIBSC MLH1/MSH2 Exon Copy Number Reference Panel was used to evaluate the workflow’s ability to detect MSH2 gene deletions.
Two 50% VAF variant samples out of the multiple references included in the standard were selected for testing. These were processed together on a single flow cell using barcode 13 and 14, alongside a third standard barcoded sample LGC/SeraCare Seraseq BRCA 1/2 Exon Deletions DNA Mix, achieving average gene coverages of 46× and 49×.
Both variants were correctly identified by the workflow (Table 2).
Table 2. Detection of MSH2 Structural Variants in NIBSC Reference Panel
| Sample | Gene | Variant Type | Variant Length (bp) | Chr | Position | Expected VAF | VAF |
|---|---|---|---|---|---|---|---|
| NIBSC | MSH2 | DEL | 19,884 | 2 | 47,400,746 | 50 | 52.17 |
| NIBSC | MSH2 | DEL | 15,376 | 2 | 47,394,320 | 50 | 61.36 |
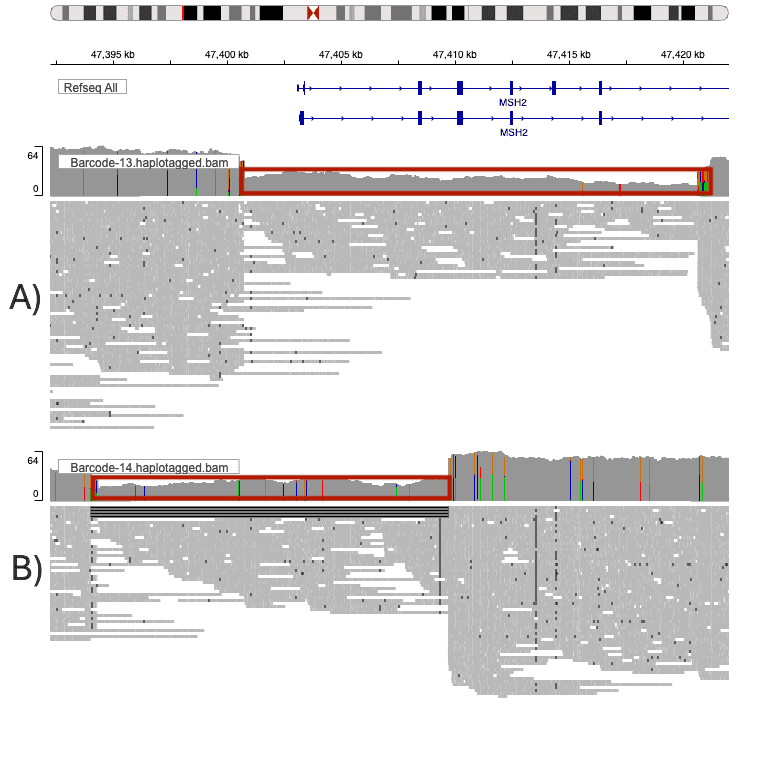
IGV visualization confirmed that the identified deletions corresponded precisely to expected exon boundaries:
- Variant A: Six-exon deletion at ~47.4 Mb on chromosome 2.
- Variant B: Two-exon deletion at ~47.39 Mb on chromosome 2.
Both events were clearly supported by soft-clipped and gapped alignments in the read data (Figure 3).
Related Materials
- Further information about the Hereditary Cancer Panel
- Store purchase
- Hereditary Cancer Panel Protocol
Tags
Share
Related Posts
Related Links
