Transcriptomic reference sets

Transcriptomic reference sets play an important role in cDNA and RNA analysis workflows such as wf-transcriptomes. These curated annotated references define gene and transcript boundaries, splice variants, and exon usage. Using a consistent, standard reference improves alignment accuracy, quantification, and reproducibility across studies.
NCBI RefSeq, GENCODE, and Ensembl provide commonly used reference sets but you may not be aware of the differences. In this post, we’ll break down these key reference resources, explore their differences, help you decide which one to use, and highlight common pitfalls to watch out for.
NCBI RefSeq
Launched in the early 2000s, the RefSeq project by the National Center for Biotechnology Information (NCBI) based in the USA, has since become one of the most widely used resources in genomics and transcriptomics.
It provides a non-redundant, high-quality reference for genomic DNA, transcripts (mRNA), and proteins for a wide range of organisms, generated through a combination of automated computational pipelines and manual expert curation. Researchers can contribute annotations to the database, but all submissions undergo rigorous review, either manually by expert curators or via automated quality control pipelines.
Ensembl
Launched in 1999 during the era of the Human Genome Project, Ensembl was developed by the European Bioinformatics Institute (EMBL-EBI) to deliver a comprehensive, evidence driven gene annotation system that could scale with the growing number of sequenced genomes and is also one of the most prominent resources.
While Ensembl does not accept direct community submissions, it collaborates extensively with large consortia and integrates third-party datasets as part of its release process. Ensembl relies heavily on its automated annotation pipelines, integrating evidence from cDNA and EST alignments, protein homology, RNA-seq data and Ab initio predictions. The result is a comprehensive and regularly updated reference annotation, with new releases every 2–3 months for a wide range of vertebrate and eukaryotic species.
GENCODE
GENCODE is a joint project between EMBL-EBI and the Wellcome Sanger Institute to provide a comprehensive and high-quality reference annotation of the human and mouse genomes, with an emphasis on manual curation. It is widely regarded as the gold standard for use in major genomics projects such as ENCODE, GTEx, and the 1000 Genomes Project.
You might notice that Ensembl and GENCODE annotations are biologically identical, since Ensembl incorporates GENCODE gene models directly. Unlike Ensembl’s frequent release cycle, GENCODE releases are less frequent but each release reflects a carefully curated and reviewed dataset, prioritising accuracy over speed.
MANE
As you can imagine, the existence of two very prominent reference sets can be problematic. MANE (Matched Annotation from NCBI and EMBL-EBI) is a collaborative project started in 2018 between NCBI RefSeq and Ensembl/GENCODE to produce a single, consistent transcript annotation per protein-coding gene in the human genome. Projects like ClinVar, HGNC, and variant annotation tools are increasingly referencing MANE.
Other
You may also encounter GenBank, NCBI’s archival sequence database. Unlike RefSeq, which is curated and non-redundant, GenBank contains all submitted sequences including raw and redundant entries. While invaluable as a record of everything deposited, GenBank is not typically used as a source for reference sets in transcriptomics workflows.
Other resources like the UCSC Genome Browser also provide downloadable gene annotations and repackage tracks from RefSeq and GENCODE, but are more commonly used for visualization and custom data integration than as primary reference sets.
Reference IDs
Here is table to demonstrate how IDs differ between the main databases using the example of TP53.
| Category | NCBI RefSeq | Ensembl / GENCODE | MANE Select |
|---|---|---|---|
| Gene symbol | TP53 | TP53 | TP53 |
| Gene ID | GeneID:7157 | ENSG00000141510 | GeneID:7157 / ENSG00000141510 |
| Transcript ID | NM_000546.6 | ENST00000269305.9 | NM_000546.6 / ENST00000269305.9 |
| Protein ID | NP_000537.3 | ENSP00000269305.4 | NP_000537.3 / ENSP00000269305.4 |
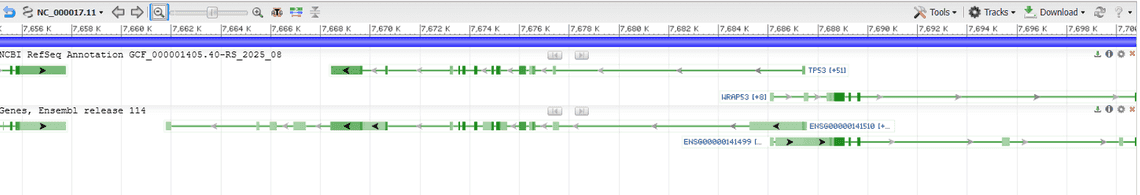
Also be aware that NCBI typically uses a numerical system to denote chromosomes eg. 1,2 whereas Ensembl prefixes chr eg. chr1, chr2.
What to use
Choosing the right reference set depends on the type of analysis being done and downstream tools. If working in a clinical context such as variant interpretation or diagnostic pipelines, RefSeq is commonly selected due to its stability and integration with databases like ClinVar. For even greater consistency between RefSeq and Ensembl, the MANE project provides a harmonized transcript per gene that is ideal for standardized reporting. For human or mouse RNA-seq studies, GENCODE is frequently favored for its comprehensive isoform coverage and Ensembl is often preferred for exploratory data analysis.
There may be cases where a dataset for an organism does not exist in both which will influence the choice. Additionally, the downstream tools or comparisons you plan to use might require one or the other. Whatever you choose, consistency throughout your analysis pipeline is key.
Common pitfalls to look out for
Here are some common issues to avoid and watch out for when working with transcriptomic reference datasets:
- Mixing reference versions, for example using GENCODE v39 for alignment and v36 for quantification, can lead to subtle mismatches and errors.
- Be sure to check that your annotation and FASTA files are aligned to the same genome build (e.g. GRCh38 or GRCh37). Mixing builds between alignment, annotation, or quantification steps can lead to mismatched coordinates and unreliable results.
- It’s easy to assume that RefSeq and Ensembl IDs map directly, but they often point to different transcript models.
- Annotation file formats can differ: GTF is the older version 2 and provided for use with legacy tools, whereas GFF is the latest with better standardisation and should be preferred where possible. Some analysis tools require one format or the other. Check the specifications
- If you’re packaging or redistributing reference sets as part of a tool, pipeline, or shared resource, make sure to check the licensing terms for each source. While most datasets (like RefSeq, Ensembl, and GENCODE) are openly available, some third-party repackaged versions (e.g. via UCSC) may have additional conditions.
Conclusion
Reference sets are foundational to any transcriptomic or genomic analysis, but navigating the options can be confusing. Whether you’re working with RefSeq, Ensembl, GENCODE, or MANE, understanding how these resources differ and where they overlap can help you make informed decisions that align with your project goals.
Table Of Contents
Related Posts
Related Links

